Advanced Cell Classifier is a data analyzer program for High Content Screening experiment to more accurately identify different phenotypes. The biggest
aim is to reduce user interaction but still preserve accuracy. The program is developed in MatLab, therefore it is platform independent.
System requirements
- MatLab 7.5 (R2007)
- Image processing toolbox
- Neural Networks toolbox
- Matlab report generator
System properties
- 96 and 384 plate format
- html report generation
- several classification methods
- quick image prediction
- versatile image visualization
Available classification methods
- LibSVM
- Neural Network
- Logistic.Weka
- MultilayerPerceptron.Weka
- RBFNetwork.Weka
- SimpleLogistic.Weka
- SMO.Weka
- RandomCommittee.Weka
- RandomForest.Weka
- BayesNet.Weka
News
ACC workshop in Lausanne,
We would like to invite you to the second Advanced Cell Classifier (ACC) and High-Content Data Chain (HCDC) workshop, in Lausanne EPFL 29. April 2010; (location will be announced).
Aim:
Give a deeper insight into advanced-level high-content screening and data analysis and practical help to install and use the software being developed in the ETH Light Microscopy Centre/RISC.
Program:
Global session
10:00-10.45: Advanced Cell Classifier, machine learning techniques, and statistical analysis of HC screens by Peter Horvath
10:45-11.30: High-Content Data Chain introduction by Karol Kozak
11.30-12.00: Pitfalls and screening in practice by Andreas Vonderheit
12.00-13.30: Lunch break
Session 1. (Data analysis track)
13.30-14.15: HCDC install and usage
14:15-15.00: ACC install and case study
Session 2. (Wet-work/general track)
13.30-13.55: Image based screening activities at EPFL by Gerardo Turcatti
13.55-14.20: Image based screening activities at ETH Zurich by Gabor Csucs
14.20-15.00: To be announced
Semi-supervised learning module
Semi-supervised learning (SSL) is a method combining the opportunities of both supervised and unsupervised learning, using unlabeled data and some supervision information to better estimate classes than unsupervised learning but requiring less field expertise than in case of supervised learning. Our primary goal is to develop a module where the user defines the positive, negative and if other known controls are available than those, and running SSL clustering we try to homogenize classes and learn them. After the first trials we conclude that using the proposed method we were able to identify ~95% of the hits with less than 10 mouse click instead of several hours teaching of supervised classifie
First genome-wide screen with ACC
We finished the primary analysis of a human genome-wide (~22.000 genes, using 4 oligos) screen. The result shows the reliability of ACC, the achieved Z' factor is 0.755, which is considered as excellent. Technical details: more than 2 million fluorescent images; ~80 million cells were classified; 92% classification accuracy measured by cross validation.
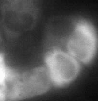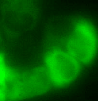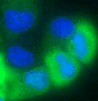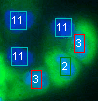
Collaboration and Masters thesis
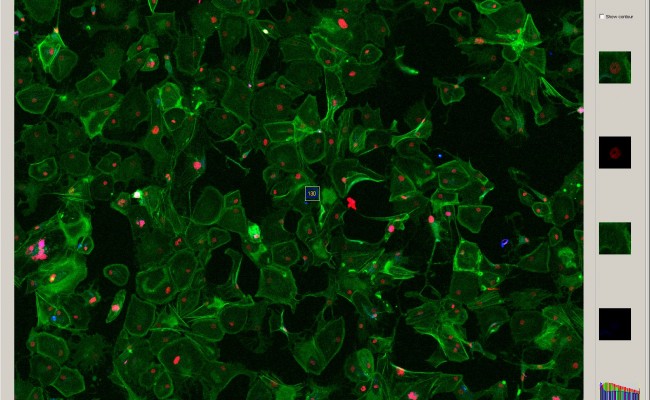
Research Institutes and Masters students are welcomed to collaborate with us to further develop or customize Advanced Cell Classifier.
For Masters students the following topics are suggested but original ideas are welcomed:
- Semi-supervised learning methods in high-content screening
The aim of this project would be to develop and implement a new classification method and compare its accuracy to supervised methods on our large screen databases
Background: programming skills and interest in bioinformatics (ETH students preferred, externals with own scholarship)
- Statistical analysis and hit detection of cell-based screens
The primary goal of this project is to better understand and detect interesting (unusual) events in cell-based high-content screens. The candidate should design a solid statistical approach for normalization and hit detection and implement it.
Background: strong statistical and mathematical, basic programming knowledge required with interest in bioinformatics (ETH students preferred, externals with own scholarship)
For research institutes:
- Joint projects in HCA, machine learning or higher-level statistical analysis
- Customization of ACC bringing new features
Cell CULTURE
Chicken embryo extract will add nutrients to your culture medium without the risk of BSA and FBS prions.
Find more information here.